I was revisiting the slides of a talk I gave a while back about graph neural networks and the material and references still hold up, so I've posted them on my personal web site:
Recent searches
Search options
#graphneuralnetworks
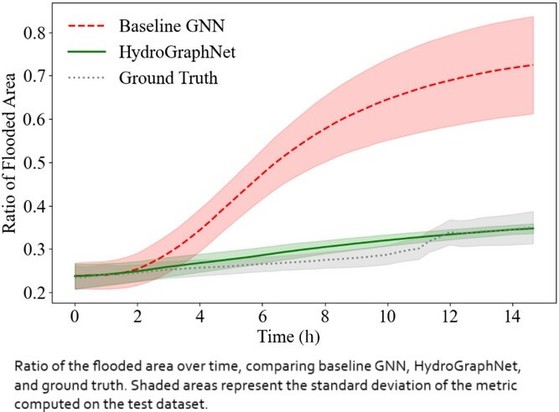
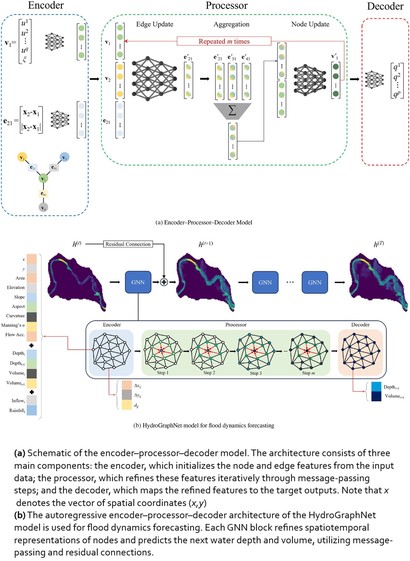
Interpretable Physics-Informed Graph Neural Networks For Flood Forecasting
--
https://doi.org/10.1111/mice.13484 <-- shared paper
--
https://github.com/MehdiTaghizadehUVa/modulus/tree/main/examples/weather/flood_modeling/hydrographnet <-- shared GitHub repository
--
#GIS #spatial #mapping #FloodForecasting #GraphNeuralNetworks #PhysicsInformedAI #Hydrology #ClimateResilience #ScientificMachineLearning #CivilEngineering #HydroGraphNet #PhyiscsNeMo #hydrology #water #flood #flooding #spatialanalysis #spatiotemporal #climatechange #model #modeling #machinelearning #AI #extremeweather #prediction #forecasting #planning #humanimpacts #loss #lossoflife #publicsafety #cost #economics #infrastructure #risk #hazard #disaster #mitigation #preparedness #naturaldisaster #HydroGraphNet #floodforecasting #dynamics #remotesensing #earthobservation
Ready for AI to crack the RNA-disease code?
GL4SDA: Predicting snoRNA-disease associations using GNNs and LLM embeddings. Computational and Structural Biotechnology Journal, DOI: https://doi.org/10.1016/j.csbj.2025.03.014
CSBJ: https://www.csbj.org/
Revolutionizing Event Detection: The DASR Framework Combats Catastrophic Forgetting
In the ever-evolving landscape of natural language processing, the DASR framework emerges as a game changer for event detection. By leveraging pre-trained language models and innovative incremental le...
An introduction to graph neural networks, with a section on "where to find them", that does not mention at all neural circuits or connectomes. WTF.
Very happy to announce our new paper accepted in @eswc_conf
#ESWC2024: "Treat Different Negatives Differently: Enriching Loss Functions with Domain and Range Constraints for Link Prediction"!
https://arxiv.org/pdf/2303.00286.pdf
w/ N. Hubert, A. Brun, and D. Monticolo
Newsletter
Dive into the world of AI security! Discover essential strategies to shield your generative AI apps from prompt injection risks. Stay ahead in the AI safety game!
#GenerativeAI #LLMs #PromptInjection #Cybersecurity #EthicalAI #ResponsibleAI #AISafety #AnomalyDetection #WeatherForecasting #GraphNeuralNetworks #DeepLearning #ClimateChange
https://gradientflow.substack.com/p/securing-ai-addressing-the-emerging
Newsletter
Dive into the world of AI security! Discover essential strategies to shield your generative AI apps from prompt injection risks. Stay ahead in the AI safety game!
#GenerativeAI #LLMs #PromptInjection #Cybersecurity #EthicalAI #ResponsibleAI #AISafety #AnomalyDetection #WeatherForecasting #GraphNeuralNetworks #DeepLearning #ClimateChange
https://gradientflow.substack.com/p/securing-ai-addressing-the-emerging
Newsletter
Dive into the world of AI security! Discover essential strategies to shield your generative AI apps from prompt injection risks. Stay ahead in the AI safety game!
#GenerativeAI #LLMs #PromptInjection #Cybersecurity #EthicalAI #ResponsibleAI #AISafety #AnomalyDetection #WeatherForecasting #GraphNeuralNetworks #DeepLearning #ClimateChange
https://gradientflow.substack.com/p/securing-ai-addressing-the-emerging
Newsletter
Dive into the world of AI security! Discover essential strategies to shield your generative AI apps from prompt injection risks. Stay ahead in the AI safety game!
#GenerativeAI #LLMs #PromptInjection #Cybersecurity #EthicalAI #ResponsibleAI #AISafety #AnomalyDetection #WeatherForecasting #GraphNeuralNetworks #DeepLearning #ClimateChange
https://gradientflow.substack.com/p/securing-ai-addressing-the-emerging
Newsletter
Dive into the world of AI security! Discover essential strategies to shield your generative AI apps from prompt injection risks. Stay ahead in the AI safety game!
#GenerativeAI #LLMs #PromptInjection #Cybersecurity #EthicalAI #ResponsibleAI #AISafety #AnomalyDetection #WeatherForecasting #GraphNeuralNetworks #DeepLearning #ClimateChange
https://gradientflow.substack.com/p/securing-ai-addressing-the-emerging
Interpretable Graph Neural Networks for Tabular Data
https://arxiv.org/abs/2308.08945
Discussion: https://news.ycombinator.com/item?id=37269376
* GNN essentially deep NN black-box models
* IGNNet: Interpretable Graph Neural Network for tab data
* notable HN comment, resp. to critique: " Right, the significance of orig. article & related research is ChatGPT-like models don't handle tabular data well & there's need for things that do"
Addendae 4
Graph Structure f. Point Clouds: Geometric Attention All You Need
https://arxiv.org/abs/2307.16662
* partly intersect my interest in knowledge graphs, fully-connected networks, and transformers in NLP and ML
* length of that discussion - though brief - exceeds the Mastodon 500-char limit, so I moved that discussion here:
Fully-connected graphs: Graph neural networks, transformers
https://persagen.com/docs/gnn-transformers.html
Hierarchical GNNs for Large Graph Generation
https://arxiv.org/abs/2306.11412
Large graphs are present in a variety of domains, including social networks, civil infrastructure, & the physical sciences. ... Graph generation is similarly widespread, with applications in drug discovery, network analysis & synthetic datasets among others. While GNN (Graph Neural Network) models have been applied in these domains their high in-memory costs restrict them to small graphs. ...
"CI-GNN: A Granger Causality-Inspired Graph Neural Network for Interpretable Brain Network-Based Psychiatric Diagnosis"
https://arxiv.org/abs/2301.01642
ROLAND: A new framework to repurpose static GNNs to dynamic GNNs.
In real life, interaction graphs are often dynamic: edges and nodes change with time
ROLAND ia built with PyG @PyTorch GraphGym to efficiently explore the GNN design space.
"On the Ability of Graph Neural Networks to Model Interactions Between Vertices"
https://arxiv.org/abs/2211.16494
Hi all,
I'm a PhD student in #MachineLearning at the technical university of Munich #TUM. I'm currently working on machine learning on graphs and machine learning-driven computional chemistry.
#ml #GraphNeuralNetworks #GNNs #compchem