Pipeline release! nf-core/ampliseq v2.13.0 - Ampliseq Version 2.13.0!
Please see the changelog: https://github.com/nf-core/ampliseq/releases/tag/2.13.0
Pipeline release! nf-core/ampliseq v2.13.0 - Ampliseq Version 2.13.0!
Please see the changelog: https://github.com/nf-core/ampliseq/releases/tag/2.13.0

Une offre de post-doc de 1 an renouvelable 1 an :
> Finaliser le développement d’une méthode d’identification moléculaire du phytoplancton utilisant le séquençage ADN haut-débit #metabarcoding
> CDD post-doc de 1 an, reconductible 1 an : démarrage à partir de mai 2025, Lieu : UMR Carrtel, Thonon https://fr-carrtel.lyon-grenoble.hub.inrae.fr/
> Projet pluriannuel #OFB et le projet européen #eDNAquaPlan
> candidature : frederic.rimet@inrae.fr
> plus de détails : https://filesender.renater.fr/?s=download&token=81a4c9dc-5e0f-49eb-a5bc-d83b53abb14d

New paper in Molecular Ecology Resources sharing data in @gbif:
Navigating Past #Oceans: Comparing #Metabarcoding and #Metagenomics of #Marine Ancient Sediment Environmental #DNA
#OpenAccess: ️
Researchers partner with #IndigenousKnowledge holders to protect threatened great desert skink Tjakuṟa https://phys.org/news/2025-01-partner-indigenous-knowledge-holders-threatened.html
#Metabarcoding clarifies the diet of the elusive and vulnerable Australian #tjakura (Great Desert #Skink, Liopholis kintorei) https://www.frontiersin.org/journals/ecology-and-evolution/articles/10.3389/fevo.2024.1354138/full
"Tjakuṟa is a large #lizard that lives in multi-tunneled communal burrows in #Australia... scientists rely on #Aṉangu Traditional Knowledge to locate Tjakuṟa burrows, estimate burrow occupancy, identify predator tracks"
The preliminary program for the AquaEcOmics symposium is now available: https://aquaecomics.symposium.inrae.fr/program2
#eDNA #omics #metagenomics #metabarcoding #dPCR #marine #freshwater #ecology #biomonitoring #aquaecomics2025
Make sure not to miss out and register before the 31th of January: https://aquaecomics.symposium.inrae.fr/
Pleased to see this published (and open access) - I’ve been contributing to this #ForestResearch / #HuttonInstitute led project for over 5 years (with most of the co-authors even longer):
Green et al (2025) The prevalence of Phytophthora in British plant nurseries; high-risk hosts and substrates and opportunities to implement best practice https://doi.org/10.1111/ppa.14044
https://mastodon.social/@aquaecomics/113837658626432987
aquaecomics - Happy to announce the program of AquaEcOmics meeting !!!
#omics #eDNA #metabarcoding #genomics #evian #nature #freshwater #marine
Access the program here:
https://aquaecomics.symposium.inrae.fr/program2

Developing and testing a new Ecological Quality Status index based on marine nematode metabarcoding: A proof of concept
https://www.sciencedirect.com/science/article/pii/S004565352402900X
ITSxpress version 2: software to rapidly trim internal transcribed spacer sequences with quality scores for amplicon sequencing
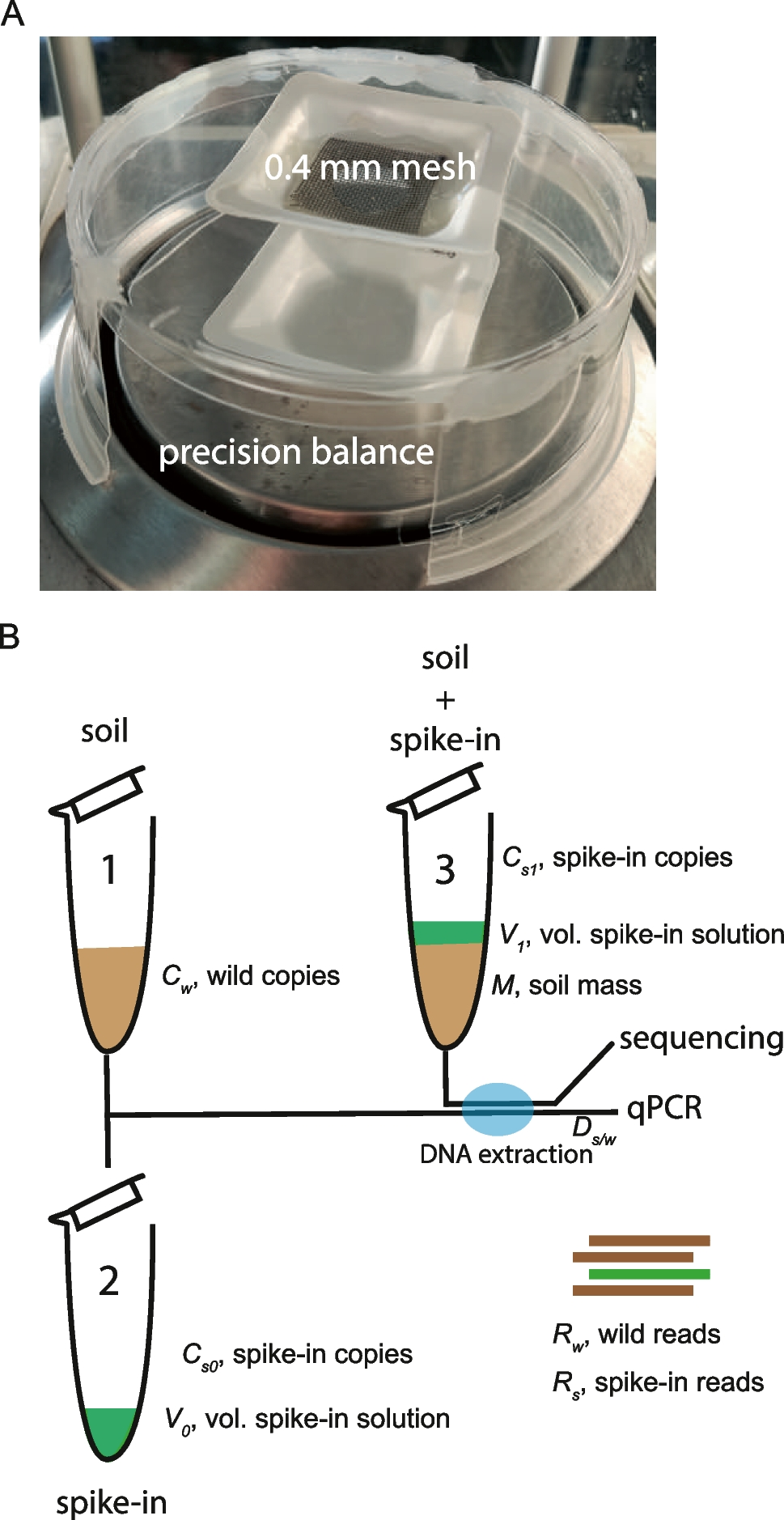
A new spike-in-based method for quantitative metabarcoding of soil fungi and bacteria
https://link.springer.com/article/10.1007/s10123-023-00422-5
Took a while, but my largest ever upload of data to the #ENA / #SRA is live, nearly 4000 #metabarcoding amplicon sequencing runs using a #Phytophthora ITS1 marker: https://www.ebi.ac.uk/ena/browser/view/PRJEB76241
Managing sequencing #metadata is hard!
Pipeline release! nf-core/ampliseq v2.12.0 - Ampliseq Version 2.12.0!
Please see the changelog: https://github.com/nf-core/ampliseq/releases/tag/2.12.0
Pipeline release! nf-core/detaxizer v1.1.0 - detaxizer 1.1.0 - Kombjuudr!
Please see the changelog: https://github.com/nf-core/detaxizer/releases/tag/1.1.0
Saving the #bats: Researchers find #bacteria and #fungi on #bat wings that could help fight deadly white-nose syndrome https://phys.org/news/2024-10-bacteria-fungi-wings-deadly-white.html
DNA #metabarcoding analyses reveal fine-scale #microbiome structures on Western Canadian bat wings https://journals.asm.org/doi/10.1128/spectrum.00376-24
"Healthy wings are critical for the survival and reproduction of bats and the wing microbiome is believed to play a major role in their susceptibility to #WhiteNoseSyndrome"
Register for the upcoming AquaEcOmics Symposium and come listen to great keynote speakers who will discuss the use of environmental omics for microbial network analyses, assessing functional diversity and evolutionary diversification and gaining insights into spatial and temporal community dynamics. https://aquaecomics.symposium.inrae.fr/ #aquaecomics2025 #eDNA #omics #metagenomics #metabarcoding #dPCR #marine #freshwater #ecology #biomonitoring
New study reveals the crucial role of #jellyfish in Greenlandic waters https://www.rcinet.ca/eye-on-the-arctic/2024/08/27/new-study-reveals-the-crucial-role-of-jellyfish-in-greenlandic-waters/
A belly full of jelly? DNA #metabarcoding shows evidence for gelatinous #zooplankton predation by several #fish species in #Greenland waters https://royalsocietypublishing.org/doi/10.1098/rsos.240797
"Scientists have commonly regarded jellyfish as a secondary food source for predatory fish, due to their low energy density and nutritional value. But... fish consume jellyfish much more frequently than previously thought."
Interested in #nextflow & anything 'meta-' ?(#metagenomics, #metabarcoding, #metatranscriptomics, #metaproteomics, #microbes #MicrobialEcology & more!)
Join us next Tuesday 13:00 CEST (3rd September) for an nf-core #bytesize for the #metaomics #nfcore special interest group! (Also on YouTube after!)
We will introduce how we want the community to work together with #users and #bioinformatics developers to make the best pipelines for anything 'meta-'
Pipeline release! nf-core/ampliseq v2.11.0 - Ampliseq Version 2.11.0!
Please see the changelog: https://github.com/nf-core/ampliseq/releases/tag/2.11.0
#16s #18s #amplicon-sequencing #edna #illumina #iontorrent #its #metabarcoding #metagenomics #microbiome #pacbio #qiime2 #rrna #taxonomic-classification #taxonomic-profiling #nfcore #openscience #nextflow #bioinformatics
AquaEcOmics - Registrations for the 2025 AquaEcOmics Symposium are now open! Register now to learn and discuss about recent advancements and challenges of omic tools and how they can be used to address pertinent scientific and management questions in aquatic ecosystems.
https://aquaecomics.symposium.inrae.fr/
#aquaecomics2025 #eDNA #omics #metagenomics #metabarcoding #dPCR #marine #freshwater #ecology #biomonitoring #metabolomics