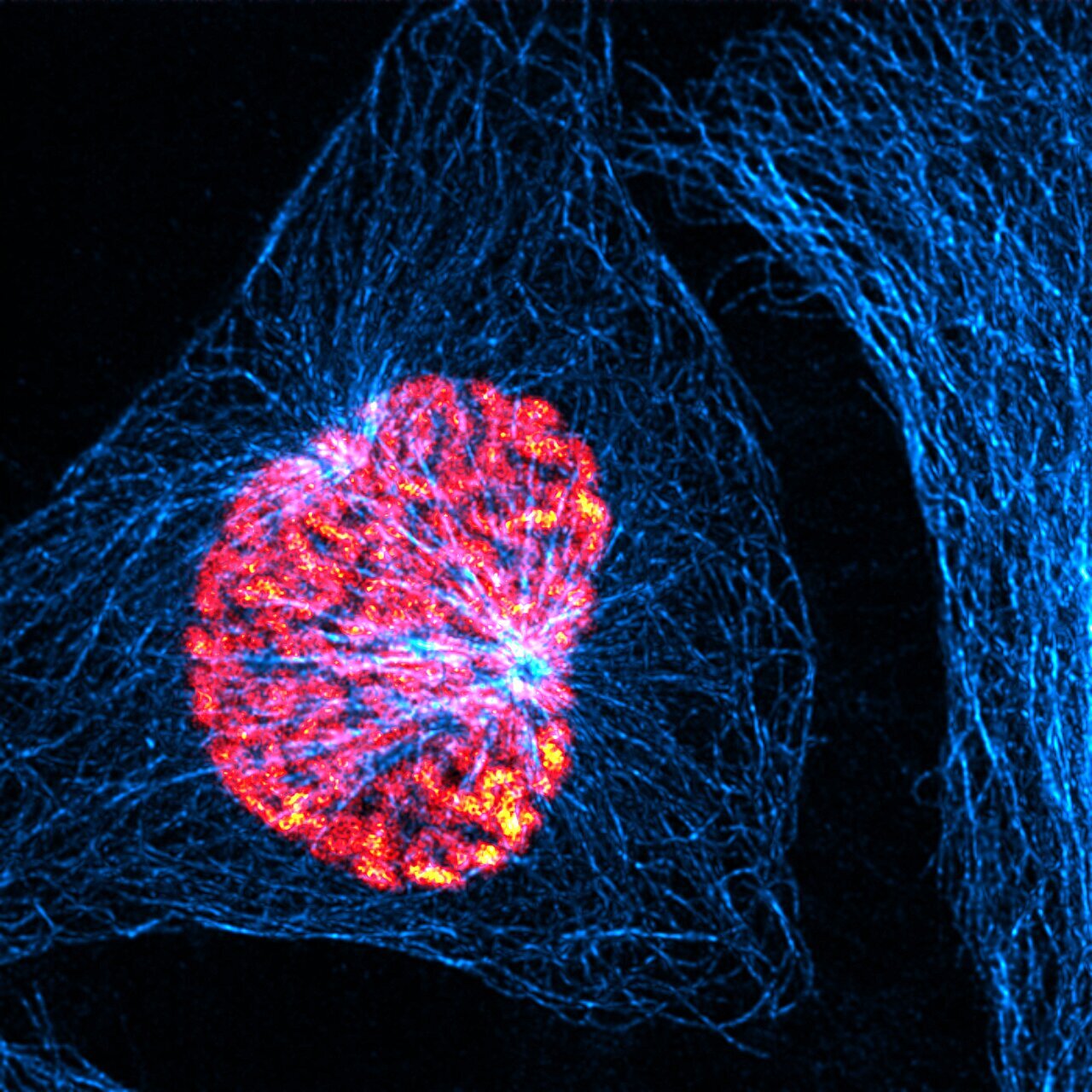
"Regulation of microtubule growth rates and their impact on chromosomal instability"
https://doi.org/doi:10.1080/15384101.2025.2485842
https://pubmed.ncbi.nlm.nih.gov/40260826/
#CellDivision #Microtubule #Cell
Recent searches
Search options
#CellDivision
"Titin cleavage in living cardiomyocytes induces sarcomere disassembly but does not trigger cell proliferation"
https://www.biorxiv.org/content/10.1101/2025.04.22.645658v1?rss=1 #CellDivision #Cell
"Cell cycle control of cohesion establishment"
https://www.biorxiv.org/content/10.1101/2025.04.18.649605v1?rss=1 #CellDivision #Cell
"The Magnitude of Telomere Shortening per Cell Division In Vivo: Implications for Lifelong Hematopoiesis in Humans"
https://www.biorxiv.org/content/10.1101/2025.04.16.649166v1?rss=1 #CellDivision #Cell
"PLK4: Master Regulator of Centriole Duplication and Its Therapeutic Potential"
https://doi.org/doi:10.1002/cm.22031
https://pubmed.ncbi.nlm.nih.gov/40257113/
#CellDivision #Microtubule
"Disruption of spindle orientation and protein localization during asymmetric cleavage by pharmacological inhibition of serotonin signaling"
https://www.biorxiv.org/content/10.1101/2025.04.19.649612v1?rss=1 #CellDivision #Cell
"Oncogene Silencing via ecDNA Micronucleation"
https://www.biorxiv.org/content/10.1101/2025.04.15.648906v1?rss=1 #CellDivision #Cell
"Mis-localization of PBPs in Staphylococcus aureus gdpP mutant contributes to β-lactam resistance and surface protein cross-wall trafficking"
https://www.biorxiv.org/content/10.1101/2025.04.15.649054v1?rss=1 #CellDivision #Cell
"Boundary Effects in Biological Planar Networks: Pentagons Dominate Marginal Cells"
https://arxiv.org/abs/2503.18855 #CellDivision #Q-Bio.Ot #Cell

"Optimal division asymmetry in E. coli bacteria"
https://www.biorxiv.org/content/10.1101/2025.04.10.648155v1?rss=1 #CellDivision #Cell
"Specific mitotic events drive left-right organizer development"
https://doi.org/doi:10.1242/dev.204687
https://pubmed.ncbi.nlm.nih.gov/40230260/
#CellDivision #Cell
"An Essential Adaptor for Apicoplast Fission and Inheritance in Malaria Parasites"
https://www.biorxiv.org/content/10.1101/2025.04.12.648511v1?rss=1 #CellDivision #Cell
"Secreted LysM proteins are required for niche competition and full virulence in Pseudomonas savastanoi during host plant infection"
https://www.biorxiv.org/content/10.1101/2025.04.11.648327v1?rss=1 #CellDivision #Cell

"A Morpho-Proteomic Atlas of Mitosis at Sub-Minute Resolution"
https://www.biorxiv.org/content/10.1101/2025.04.09.647158v1?rss=1 #CellDivision #Mitosis #Cell

"Atomic Conformational Dynamics and Actin-Crosslinking Function of Alpha-Actinin Revealed by SimHS-AFMfit"
https://www.biorxiv.org/content/10.1101/2025.04.06.647477v1?rss=1 #CellDivision #Dynamics #Actin

"Proximity-based activation of AURORA A by MPS1 potentiates error correction"
https://doi.org/doi:10.1016/j.cub.2025.03.018
https://pubmed.ncbi.nlm.nih.gov/40203828/
#CellDivision #Cell
"A local composition of peptidoglycan drives the division site selection by MapZ in Streptococcus pneumoniae"
https://www.biorxiv.org/content/10.1101/2025.04.08.647842v1?rss=1 #CellDivision #Cell

"Aurora Kinase A and B inhibition abrogates 'Neosis', a non-mitotic cell division of GBM residual cells and prevents GBM recurrence"
https://doi.org/doi:10.1038/s41388-025-03372-6
https://pubmed.ncbi.nlm.nih.gov/40195468/
#CellDivision #Cell
"In vivo regulation of an endogenously-tagged protein by a light-regulated kinase"
https://doi.org/doi:10.1093/g3journal/jkaf073
https://pubmed.ncbi.nlm.nih.gov/40194518/
#CellDivision #Cell
About 100 cells divide every second in our body. A key protein in cell division is a protein kinase termed Plk1, because it activates other proteins involved in this process. Plk1 is also overexpressed in many types of cancer. This makes it a promising target for cancer therapies. However, drugs that inhibit Plk1 have often proven ineffective.
https://phys.org/news/2025-04-cell-division-molecular-cells.html